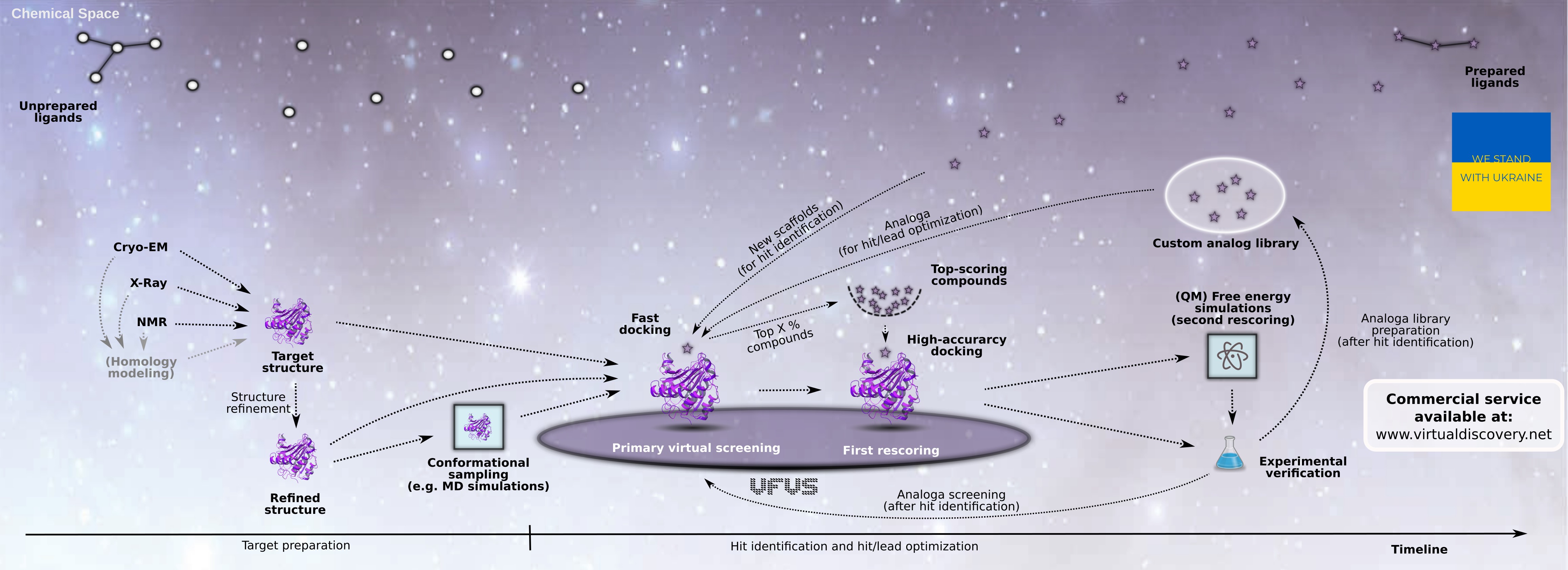
Introduction
VirtualFlow is a versatile, parallel workflow platform for carrying out virtual screening related tasks on Linux-based computer clusters of any type and size which are managed by a batchsystem (such as SLURM).
Currently, there exist two versions of VirtualFlow, which are tailored to different types of tasks:
They use the same core technology regarding the workflow management and parallelization, and they can be used individually or in concert with each other. Additional versions are expected to arrive in the future.
Pre-built ready-to-dock ligand libraries for VFVS are available for free (in the download section).
How to cite: If you are using VirtualFlow, please cite the following papers in relevant publications:
- Christoph Gorgulla, Andras Boeszoermenyi, Zi-Fu Wang, Patrick D. Fischer, Paul W. Coote, Krishna M. Padmanabha Das, Yehor S. Malets, Dmytro S. Radchenko, Yurii S. Moroz, David A. Scott, Konstantin Fackeldey, Moritz Hoffmann, Iryna Iavniuk, Gerhard Wagner, Haribabu Arthanari An open-source drug discovery platform enables ultra-large virtual screens. Nature 580, 663–668 (2020). https://doi.org/10.1038/s41586-020-2117-z
- Gorgulla, C., Fackeldey, K., Wagner, G., & Arthanari, H. (2020). Accounting of Receptor Flexibility in Ultra-Large Virtual Screens with VirtualFlow Using a Grey Wolf Optimization Method. Supercomputing Frontiers and Innovations, 7(3), 4–12. https://doi.org/10.14529/jsfi200301
- Gorgulla, C., Çınaroğlu, S.S., Fischer, P.D., Fackeldey, K., Wagner, G. and Arthanari, H., 2021. VirtualFlow Ants—Ultra-Large Virtual Screenings with Artificial Intelligence Driven Docking Algorithm Based on Ant Colony Optimization. International Journal of Molecular Sciences, 22(11), p.5807. https://doi.org/10.3390/ijms22115807
- Gorgulla, C., Fackeldey, K., Wagner, G. and Arthanari, H., 2020. Accounting of receptor flexibility in ultra-large virtual screens with VirtualFlow using a grey wolf optimization method. Supercomputing frontiers and innovations, 7(3), p.4. https://dx.doi.org/10.14529%2Fjsfi200301
VirtualFlow @COVID Project: https://vf4covid19.hms.harvard.edu/
Commercial Service: A commercial service for ultra-large virtual screenings based on VirtualFlow is available via the company Virtual Discovery, Inc.